Research
Aneuploidy, defined as an incorrect chromosome number or change in the chromosome content, is the most common genetic change in cancer and occurs in >90% of tumors. The most well-established driver of aneuploidy is chromosome segregation errors that occur during mitotic cell division.
The Frank Mason lab is interested in understanding how these mitotic errors arise and discovering the consequences of these errors with respect to tumorigenesis. Aneuploidy and genomic instability cause a host of transcriptional, metabolic, and proteotoxic stresses within cells that can ultimately promote transformation, metastasis, and resistance to therapies.
Our lab seeks to better understand the causes and consequences of genomic instability through three primary areas of investigation.
Genomic Instability and Intratumoral Heterogeneity in Cancer
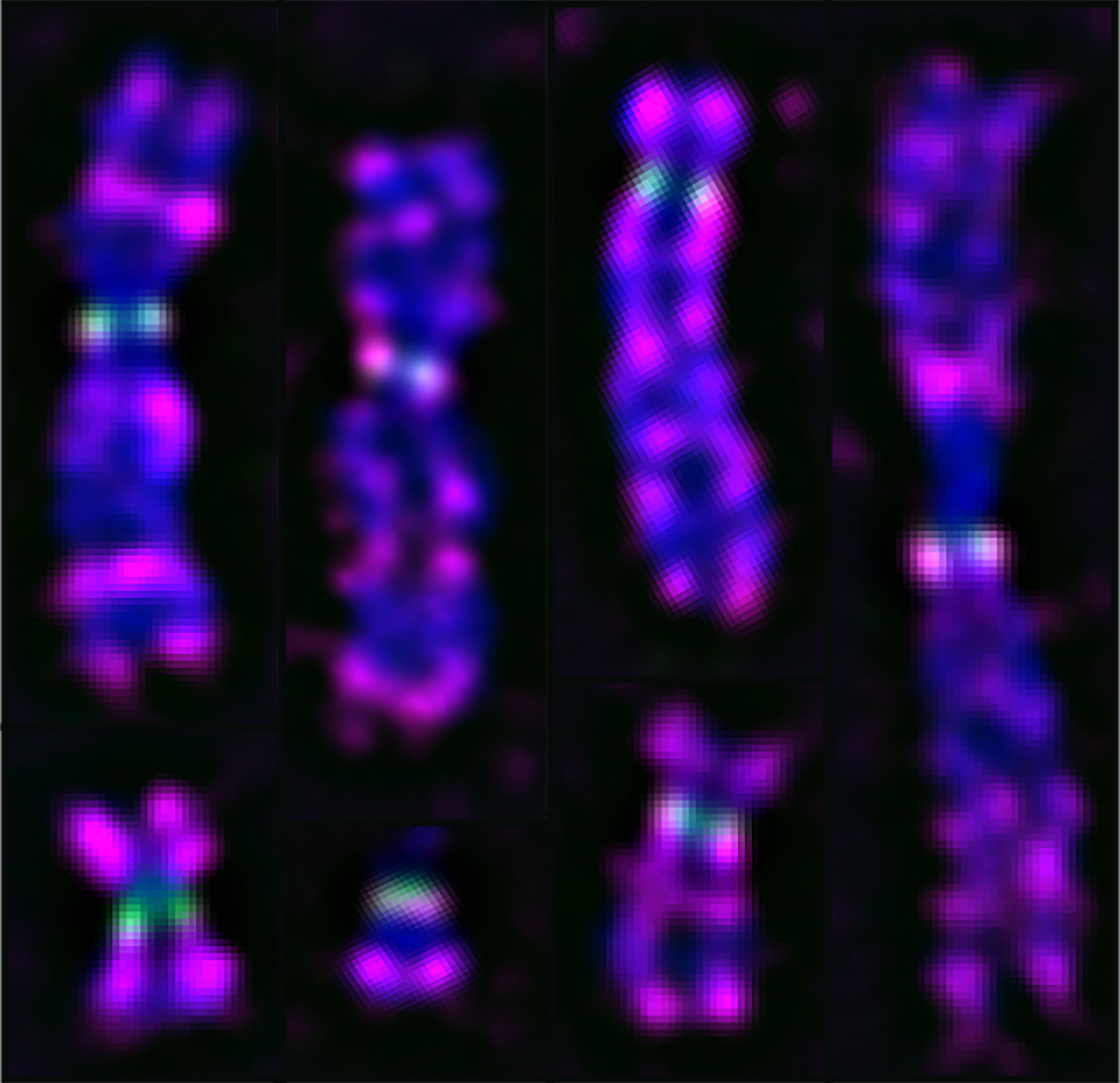
An emphasis of the lab is our study of the epigenetic regulator and tumor suppressor SETD2. SETD2 is a methyltransferase known to modify histone H3 at Lys36 and a-tubulin at Lys40 and regulates myriad aspects of the DNA damage response. SETD2 is mutated in many cancer types, including clear cell renal cell carcinoma (ccRCC) and acute lymphoblastic leukemia (ALL), and the loss of SETD2 in these tumors promotes metastasis, resistance to chemotherapeutics, and worse progression free survival.
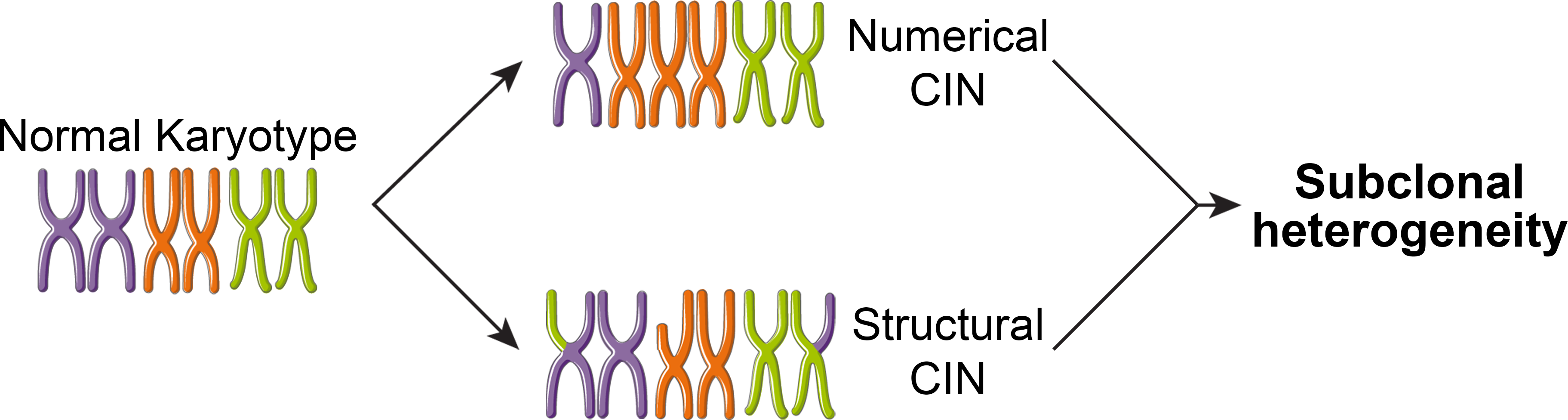
Our lab hypothesizes that these adverse outcomes are attributed to an increase in intratumoral heterogeneity associated with SETD2 loss. We have identified that SETD2 loss leads to both structural (chromosomal rearrangements) and numerical (gain or loss of whole chromosomes) chromosomal instability.
The Mason Lab is lab is interested in understanding the consequences of these different types of chromosomal instability and how these processes influence intratumoral genetic heterogeneity and other cells within the tumor microenvironment.
Regulation of Chromosome Segregation
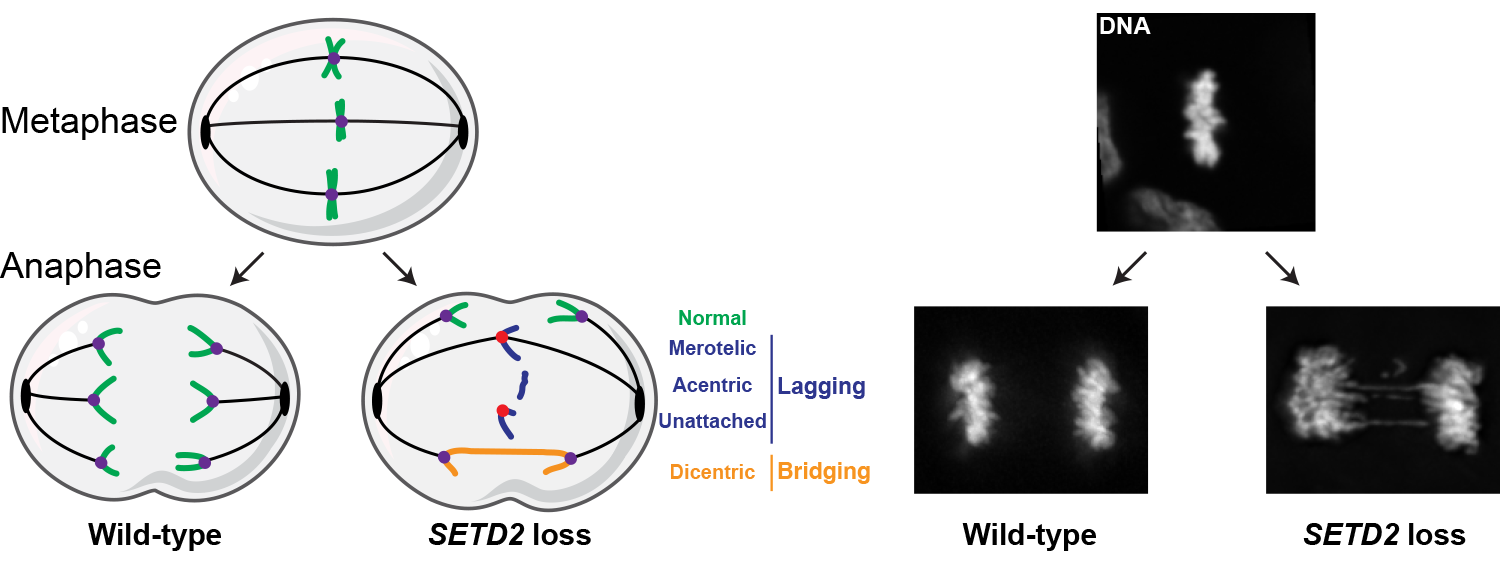
During each cell cycle, DNA is replicated then divided between the two new daughter cells such that each new cell has an accurate copy of each chromosome. To properly segregate during mitosis, cellular structures called microtubules grow and attach to each chromosome at its center, or centromere. Each chromosome has one centromere, which becomes the site for assembly of the kinetochore, the macromolecular structure connecting microtubules to centromeres. Errors during DNA replication can generate chromosomes with too many (di- or tri-centric) or no (acentric) centromeres, which may then fail to segregate properly.
The Mason Lab recently discovered that SETD2 prevents the formation of dicentric and acentric chromosomes during DNA replication. Additionally, we found that loss of SETD2 results in structural chromosome instability. For example, SETD2 deletion or inhibition causes the formation of isochromosomes, which are chromosomes with mirror-imaged symmetry. Isochromosome formation leads to simultaneous amplification of the incorporated chromosome arm and loss or deletion of the other arm.
Our lab is interested in investigating which pathway(s) regulate isochromosome formation, the loci where these inversions occur, and the cellular consequences of these chromosomal aberrations and mis-segregations.
Consequences of Epigenetic Dysregulation in Cancer
Epigenetic dysfunction is a common occurrence across many tumor types, leading to transcriptional reprogramming. While SETD2 functions as an epigenetic regulator, binding actively transcribing RNA polymerase II and mediating H3K36 trimethylation during transcription, the loss of SETD2 does not affect genic transcription.
The Mason Lab is using SETD2 as a model to understand other mechanisms by which epigenetic regulators may preserve genome maintenance and cell fitness. These potentially include epigenetic regulation of chromosome organization and stability through effects on centromeres and/or telomeres, impact on alternative splicing, and/or altering RNA-protein interactions.